Johannes Haupt remerge.io
How to put a custom pytorch module into the fastai Learner framework
Define a custom pytorch neural network module as a Learner in the fastai library to flexibly use the fastai functionality.
The problem
We have an application where we want to define our own model architecture in pytorch. I know how to define the network architecture as a nn.Module and I have created an __init__ and forward method. But I do not want to spend the time and effort to create a train method for the network to fit it to data, because the fastai library has already implemented model training and includes many helpful features like the progress bar, validation score or the learning rate finder. Can’t I put my model into the fastai training framework? Yes, with a bit of tinkering that works well!
import numpy as np
from sklearn.datasets import make_classification
X, y = make_classification(n_samples=1000, n_features=20, n_informative=10, random_state=123)
X_train, y_train = X[:500,:], y[:500]
X_valid, y_valid = X[500:,:], y[500:]
We have created a customer neural network module. The example below is a very simple network, so we could as well use the fastai tabular model. However, for my own use cases, I’ve found myself needed to implement architectures not covered by the fastai library.
import torch
import torch.nn as nn
from torch.autograd import Variable
class NNet(nn.Module):
def __init__(self, input_dim, hidden_layer_sizes, loss, sigmoid=False):
"""
input_dim : int
Number of input variables
hidden_layer_sizes : list of int
List of the number of nodes in each fully connected hidden layer.
Can be an empty list to specify no hidden layer
loss : pyTorch loss
sigmoid : boolean
Sigmoid activation on the output node?
"""
super().__init__()
self.input_dim = input_dim
self.layer_sizes = hidden_layer_sizes
self.iter = 0
# The loss function could be MSE or BCELoss depending on the problem
self.lossFct = loss
# We leave the optimizer empty for now to assign flexibly
self.optim = None
hidden_layer_sizes = [input_dim] + hidden_layer_sizes
last_layer = nn.Linear(hidden_layer_sizes[-1], 1)
self.layers =\
[nn.Sequential(nn.Linear(input_, output_), nn.ReLU())
for input_, output_ in
zip(hidden_layer_sizes, hidden_layer_sizes[1:])] +\
[last_layer]
# The output activation depends on the problem
if sigmoid:
self.layers = self.layers + [nn.Sigmoid()]
self.layers = nn.Sequential(*self.layers)
def forward(self, x):
x = self.layers(x)
return x
def fit(self, data_loader, epochs, validation_data=None):
for epoch in range(epochs):
running_loss = self._train_iteration(data_loader)
val_loss = None
if validation_data is not None:
y_hat = self(validation_data['X'])
val_loss = self.lossFct(input=y_hat, target=validation_data['y']).detach().numpy()
print('[%d] loss: %.3f | validation loss: %.3f' %
(epoch + 1, running_loss, val_loss))
else:
print('[%d] loss: %.3f' %
(epoch + 1, running_loss))
def _train_iteration(self,data_loader):
running_loss = 0.0
for i, (X,y,g) in enumerate(data_loader):
X = X.float()
y = y.unsqueeze(1).float()
X = Variable(X, requires_grad=True)
y = Variable(y)
pred = self(X)
loss = self.lossFct(pred, y)
self.optim.zero_grad()
loss.backward()
self.optim.step()
running_loss += loss.item()
return running_loss
def predict(self, X):
X = torch.Tensor(X)
return self(X).detach().numpy().squeeze()
Placing a custom torch network into the fastai Learner class
custom_nnet = NNet(X.shape[1], [10,10], loss = None)
The fastai Learner class combines a model module with a data loader on a pytorch Dataset, with the data part wrapper into the TabularDataBunch class. So we need to prepare the DataBunch (step 1) and then wrap our module and the DataBunch into a Learner object.
import fastai
import fastai.tabular
from torch.utils.data import Dataset
The easiest way to create a DataBunch without committing to the fastai framework to prepare the data is via the TabularDataBunch.create function. The create() function takes a torch Dataset as input.
class TabularData(Dataset):
def __init__(self, X, y):
"""
Torch data Loader for experimental data
X : array-like, shape (n_samples, n_features)
The input data.
y : array-like, shape (n_samples,)
The target values (class labels in classification, real numbers in regression).
g : array-like
The group indicator (e.g. 0 for control group, 1 for treatment group)
"""
self.X = X
self.y = y
def __len__(self):
return self.X.shape[0]
def __getitem__(self, idx):
return self.X[idx, :], self.y[idx]
Make sure to match the data type and module to Long/float64. If you get errors like RuntimeError: Expected object of scalar type Float but got scalar type Double for argument #4 'mat1' when calling fit, then the data type and model type don’t match.
data_fastai = fastai.tabular.TabularDataBunch.create(train_ds=TabularData(X_train.astype(np.float64),y_train.astype(np.float64)), valid_ds=TabularData(X_valid.astype(np.float64), y_valid.astype(np.float64)), bs=64)
Actually wrapping the custom module into a Learner class is straightforward. The Learner init takes as arguments the DataBunch, the pytorch module and a torch loss function.
custom_nnet = custom_nnet.double()
fastai_nnet = fastai.Learner(data=data_fastai, model=custom_nnet, loss_func=nn.MSELoss())
At this point you can start using the functionality of the fastai library for your custom model.
fastai_nnet.fit(3, 1e-5)
Total time: 00:01 <p><table style='width:300px; margin-bottom:10px'>
</table>
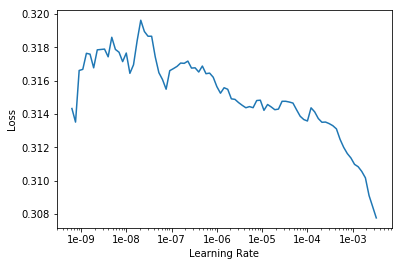
In my case, I wanted to use the learning rate finder to optimize the learning rate.
fastai_nnet.lr_find(start_lr=1e-10, end_lr=1e-2, num_it=100)
LR Finder is complete, type {learner_name}.recorder.plot() to see the graph.
fastai_nnet.recorder.plot()
That’s it! You will never have to manually implement the fit method again!